Plant breeding for climate change: Opportunities for adaptation and mitigation
Patrick F. Byrne, Department of Soil and Crop Sciences, Colorado State University, Fort Collins, Colorado USA 80523 (Patrick.Byrne@colostate.edu)
Learning Objectives
- Summarize the past, present, and future use of plant genetic resources in plant breeding.
- Provide examples of strategies and successes of employing plant genetic resources to help plants adapt to climate change.
- Explain the potential use of plant genetic resources in breeding for climate change mitigation.
outline
- Introduction
- Past, present, and future use of plant genetic resources in plant breeding
- From genebanks to farmers’ fields
- Breeding plants for adaptation to climate change
- Breeding plants for climate change mitigation
- Conclusions
- References
- Acknowledgments
1. Introduction
Genetic diversity is the foundation upon which plant breeding progress rests. Therefore, diverse genetic resources have always played a key role in the improvement of crops from wild progenitors to elite cultivars. Given the relatively narrow genetic base of most modern crops, access to a broad range of genebank resources is essential to help breeders adapt crops to the extreme conditions predicted under climate change, as well as to breed plants that can mitigate those conditions.
2. Past, Present, and Future Use of Plant genetic resources in Plant Breeding
To place this chapter in perspective, we begin with a condensed, subjective assessment of the use of plant genetic resources (PGR) by plant breeders through the past, present, and future eras of plant breeding.
Past
Crop improvement in the broad sense has a long history of altering the genetics of plant populations for the benefit of human society. Plant breeding has had numerous notable achievements from the domestication of wild plants, to selection by generations of farmers for local adaptation, to scientific breeding starting around 1900. Often this progress has been accompanied by a narrowing of genetic diversity. Modern breeders have used genebank resources occasionally, especially for disease resistance traits controlled by major genes, but not for quantitative traits like drought tolerance, with their more complex inheritance. The vulnerability of major crops due to limited genetic diversity was emphasized by the southern corn leaf blight epidemic in the U.S. in 1970-71 (Ullstrup, 1972), but relatively little progress has been made to broaden the genetic base of many crops. Further description of the history of plant breeding is contained in the eBook chapter, From Wild Species to Landraces and Cultivars.
Present
There is growing recognition of the value of PGR, especially to help crops adapt to the stresses brought about by climate change (FAO, 2015). However, identifying the most useful accessions remains a challenge, as does incorporating the beneficial variation into commercially viable cultivars. Initiatives have been completed or are underway to conduct comprehensive phenotypic and genotypic evaluations of collections to provide baseline data for their future use (Martini et al., 2021). The rapidly declining cost of genome sequencing has made it increasingly feasible to sequence entire collections, but phenotyping continues to be difficult and expensive.
Future
A more precise, data-driven use of PGR is expected in the future. Data will be integrated from multiple sources—including passport data, phenotypic and genotypic data, and knowledge of molecular biological mechanisms—to predict the most promising plant genetic resource accessions for a given trait (Martini et al., 2021). A second application of prediction is to identify the alleles to transfer from an accession to an elite cultivar background, thus facilitating the prebreeding process (Martini et al., 2021). If a specific beneficial allele is identified, then genome editing is another strategy that could be pursued (see section on Genome Editing below). The future might also see breeding programs that target climate mitigation traits, e.g., plants with deep roots that sequester carbon better than conventional crops or that demonstrate biological nitrification inhibition (Subbarao et al., 2021).
3. From genebanks to farmers’ fields
Resources in plant genebanks offer tremendous potential for improving crops for adaptation to the altered biotic and abiotic environments predicted under climate change. However, the improvement strategy can take a number of different routes. Some of the factors that will determine the most logical route for a given crop are the extent of plant genebank collections and the characterization of the crop and its wild relatives, the state of genetic and genomic resources, the level of knowledge of plant physiology or biochemistry relevant to a trait of interest, and the ease of cross-pollinating or transforming a given species. A generalized view of the path from germplasm acquisition by a genebank to farmers’ fields is shown in Figure 1.
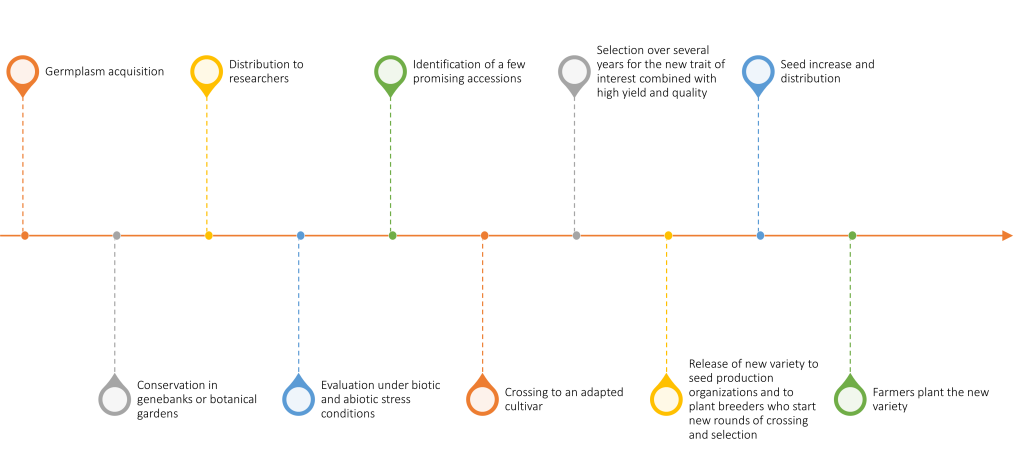
Dozens of crop species now have reference genome sequences available, but knowledge of genes controlling specific traits is still very limited. Thus, although gene editing and genetic engineering will be potential pathways for some crops and traits, more conventional breeding methods based on cross-pollination, often assisted with genomic information, will likely be the favored strategy in most cases. Access to viable, diverse, well-characterized, and accurately identified germplasm will be crucial to plant breeding progress under the uncertain conditions of the future.
4. Breeding plants for adaptation to climate change
The genetic diversity conserved in the word’s genebanks offers great promise for adapting crops to climate-induced stresses. A number of approaches, discussed in this section, have been employed to detect and harness that diversity.
4.1. Large-scale evaluations of genebank accessions
The potential utility of genebank accessions is often assessed initially through a large-scale evaluation, either of the entire crop collection or a selected subset. The huge number of accessions in some collections can be a barrier to using genebank resources effectively. To address this problem, core subsets (groups of accessions containing ~10% of the total collection of a crop while including the majority of allelic diversity) have been identified for a number of crops (Brown, 1989; Byrne et al., 2018; Vikram et al., 2021). Subsets for climate adaptation traits could also be selected based on a Focused Identification of Germplasm Subsets (FIGS) strategy (Berger et al., 2013; Mackay et al., 2007). In this strategy, accessions are identified based on climatic and other habitat characterization data from collection sites. For example, Berger et al. (2013) gave the example of selecting accessions with tolerance to drought during flowering or grain-filling based on collection site data.
Large-scale evaluations have been made more feasible through the development of high-throughput, lower-cost phenotyping technologies, as explained by Araus and Kefauver (2018) and Volk et al. (2021). Examples of evaluation for climate adaptation traits are the collection of canopy spectral reflectance data in 540 spring wheat (Triticum aestivum) accessions (Bowman et al., 2015); assessment of yield-related traits under drought stress in 590 West African sorghum (Sorghum bicolor) accessions (Faye et al., 2022); and evaluation of 486 soybean (Glycine max) accessions for high germinability of seed produced under high temperatures (Smith et al., 2008).
In addition to increasing the frequency of stressful conditions, climate change might also create new opportunities for crops in regions where they have not been grown previously. Thus, Schaffasz et al. (2019) evaluated a diverse set of 338 sorghum accessions in Central Europe for chilling tolerance, high-latitude adaptation, and bioenergy-related agronomic traits.
Video 1. “Sorghum diversity: Origins and importance” is a video produced by Colorado State University Center for Science Communication. It describes the importance of sorghum in semi-arid areas of Africa and how genetically diverse materials can be used in breeding programs to improve production traits.
Video 2. “Sorghum diversity: Breeding for temperate adaptation” is a video produced by Colorado State University Center for Science Communication. Sorghum breeding programs are developing shorter new varieties that can flower in North American photoperiodic conditions.
4.2. Breeding for biotic stress tolerance
Crop yield losses due to insect pests and pathogens are predicted to increase significantly as temperatures rise (Irish and Volk, 2023). In maize (Zea mays), for example, an average increase of 31% in production losses due to insects has been predicted if global temperature rises by 2 °C (Deutsch et al., 2018). The destructive pest fall armyworm (FAW; Spodoptera frugiperda J.E. Smith) is native to the Americas, but has expanded its range since 2016, infesting maize fields in 47 African and 23 Asian countries (Singh et al., 2021). Increasing temperatures speed up the development of FAW eggs, pupae, and larvae, foreshadowing even greater damage in the future (Huang et al., 2021; Diaz-Alvarez et al., 2021). Fortunately, resistance to FAW has been identified in a diverse array of maize landraces and other germplasm, including crop wild relatives (Singh et al., 2021). The International Maize and Wheat Improvement Center (CIMMYT) has recently developed three FAW-tolerant hybrids for eastern and southern Africa (Singh et al., 2021).

Stem rust caused by the fungus Puccinia graminis f. sp. tritici is among the most destructive diseases of wheat, and outbreaks of the disease are predicted to become more frequent as a result of climate change (Prank et al., 2019). The appearance and spread of virulent races of the pathogen have galvanized the wheat research community to find more diverse and durable sources of resistance. Recently a highly effective stem rust resistance gene was cloned from Aegilops sharonensis, a wild diploid relative of wheat. The gene was identified following genome sequencing of a line derived from Ae. sharonensis accession AS_1644, which was collected in Israel and conserved by Tel Aviv University (Yu et al., 2022; Millet et al., 2017). The gene has been inserted through genetic engineering into a susceptible wheat cultivar, resulting in lines that are resistant to all known strains of the pathogen.

Late blight caused by the oomycete Pythophthora infestans is one of the most damaging diseases of potato (Solanum tuberosum). Durable resistance in potato cultivars has been an elusive goal, but fortunately resistance has been identified in a large number of wild potato species, as reported by Blossei et al. (2022) and Karki et al. (2021). For many of these resistance sources, the responsible genes have been characterized. A challenge in using these wild species in a breeding program is that most are diploid, whereas cultivated germplasm is tetraploid. The difference in ploidy level leads to incompatibility of crosses in most cases. Blossei et al. (2022) discuss strategies for overcoming this interploidy challenge and thus utilizing the wealth of wild potato germplasm in the world’s genebanks.

4.3. Breeding for abiotic stress tolerance
Abiotic stress is predicted to increase in many of the world’s crop growing regions. Heat and drought stress have received the most attention, but other forms of abiotic stress that could become more frequent include flooding, salinity, ozone, and unseasonably cold temperatures. Important factors that affect the severity of a stress include the stress intensity and duration, the growth stage at which the stress occurs, and whether other stresses occur simultaneously.
Plant breeders have attempted to improve abiotic stress tolerance by evaluating diverse germplasm in a number of ways: using managed stress field environments, growing materials in multiple locations where variable levels of a stress are likely to occur, or imposing stress conditions in greenhouse or growth chamber environments. Crop wild relatives and landrace accessions have been evaluated frequently as potential sources of stress tolerance, though breeders are sometimes reluctant to incorporate unadapted germplasm into their elite breeding populations.

Many traits have been investigated for their potential as predictors for identifying abiotic stress tolerant germplasm. Two examples are provided below: slow wilting and root architecture traits.
Slow wilting
Slow wilting is a phenotype observed in soybean (Glycine max) and other crops whereby canopy wilting under drought stress is delayed relative to an average cultivar. In soybean, the trait was first reported in a Japanese landrace, PI 416937; that accession and an introduction from Nepal (PI 471938) have been used to develop several drought tolerant soybean varieties (Kunert and Vorster, 2020). Although the mechanisms are not completely identified, slow wilting is believed to be largely due to reduced transpiration early in the growing season that conserves moisture in the soil profile for later in the season. Enhanced exploration for soil water by the root system might also be involved in some genotypes (Ye et al., 2020). Using dynamic crop modeling, Sinclair et al. (2010) found that the slow wilting trait increased yield in nearly 80% of years across most regions of the U.S.

Root architecture traits
Although notoriously difficult to evaluate, root systems have received increasing attention as a target for enhancing crop adaptation to the abiotic stresses aggravated by climate change (Lynch, 2022; Ober et al., 2021). Improved below-ground phenotyping methods and increased knowledge of the genetic loci controlling root architecture have now made selection for root traits more feasible (Ober et al., 2021). A complicating factor is that the ideal root phenotype differs according to environmental conditions, and even within the same location those conditions are increasingly variable year to year. Deep root systems may be valuable to access stored moisture deep in the profile in a dry year, but shallower roots are better able to take advantage of intermittent rainfall throughout the growing season (Ober et al., 2021). Despite the variability and uncertainty, breeders are starting to make progress by focusing on prevailing conditions within a given target environment.
Crop wild relatives, with their need to survive in often harsh environments, have provided genetic variability for root traits that resulted in improved crop performance (Leigh et al., 2022). For example, introgression of a chromosome segment from wild emmer wheat (Triticum durum subsp. dicoccoides) into a durum wheat (T. durum) cultivar was reported to result in deeper roots and improved yield under drought stress (Merchuk-Ovnat et al., 2016). A chromosome translocation from another wheat relative, Agropyron elongatum, into bread wheat also resulted in deeper rooting under drought conditions (Placido et al., 2020). A notable example of a beneficial root trait from a landrace is the deep rooting allele of DRO1, discovered in the traditional rice (Oryza sativa) cultivar ‘Kinandang Patong’ from the Philippines, which improved rice yield under drought (Uga et al., 2013).
In addition to root morphology and behavior, research into the microbial communities associated with roots is another avenue being explored to improve climate resilience. The root microbiome has been shown to influence tolerance to drought and salinity (Kim et al., 2012; Fatima and Arora, 2019), and in some cases to differ in crop ancestors compared to the domesticated species (Schmidt et al., 2016). As knowledge of microbial-host plant interactions improves, it is possible that the microbiomes associated with genebank accessions could provide another plant breeding opportunity for climate adaptation.
4.4. PREBREEDING
Many types of PGR cannot be incorporated directly into a breeding program due to lack of adaptation to a target environment, barriers to cross-pollination, or poor end-use quality. Therefore, prebreeding is often considered a necessary first step, whether for climate adaptation or improving any other trait. According to the Crop Genebank Knowledge Base (2022), “Prebreeding refers to all activities designed to identify desirable characteristics and/or genes from unadapted materials that cannot be used directly in breeding populations and to transfer these traits to an intermediate set of materials that breeders can use.” Typically, prebreeding involves crossing wild or unadapted germplasm to an adapted material, followed by one or more backcrosses to the adapted parent, and evaluating for the traits of interest. The Crop Trust has funded 19 prebreeding projects on various crops (Crop Trust, 2022), and additional examples of prebreeding activities are provided by Byrne et al. (2018).
4.5. Genome editing
A powerful, recently developed set of techniques for altering the genome is known as genome editing or gene editing. CRISPR (or CRISPR-Cas) is the most commonly used method for editing genomes due to its specificity, flexibility, ease of use, and relatively low cost. A description of genome editing applications in plant science is provided by Zhang et al. (2017), and the Microbiology Society has produced a video explaining the origin and application of the CRISPR-Cas system.
In contrast to genetic engineering, genome editing targets very specific locations in the genome and in most cases does not result in foreign DNA in the final product. Most current applications of genome editing result in a gene ‘knock out’, i.e., rendering the gene non-functional. However, it is also possible to tweak the DNA sequence of a gene to a more favorable form or to insert (‘knock in’) a gene. Although genome editing is easier than other methods, it does require detailed sequence information for the targeted gene. If the goal is crop improvement, one would also have to know (or hypothesize) the effect of that gene on the trait of interest and potentially on other traits.
With regard to climate change, some of the areas where genome editing has been shown to hold promise include the following categories of traits (Karavolias et al., 2021), with examples provided in the indicated references:
- Drought tolerance (Shi et al., 2017, maize)
- Salinity tolerance (Zhang et al., 2019, rice)
- Disease resistance (Kumar et al., 2018, barley, Hordeum vlugare)
- Yield improvement under stress (Wang et al., 2018, wheat)
- Enhanced nutrition (Kaur et al., 2020, banana, Musa spp.)
Genome editing may have particular relevance for genes identified in plant genetic resources. Once scientists know the DNA sequence of a beneficial allele, for example in a wild relative, the corresponding gene in an adapted cultivar can be precisely edited without bringing along all the negative baggage of the wild species.
Although there is considerable enthusiasm for applying genome editing to crop improvement, there are risks and uncertainties associated with the use of the technology, including an uncertain global regulatory environment and unclear intellectual property status (Pixley et al., 2022).
De novo domestication
One strategy for developing more heat and drought tolerant cultivars using gene editing is to start with a naturally tolerant crop wild relative and domesticate the species de novo. This could be done by introducing genes associated with previous domestication events (e.g., for reduced shattering in wheat or less stem branching in maize) that improve yield, harvestability, or utilization of the crop (Zsogon et al., 2022). This approach has become more feasible due to the development and refinement of gene editing techniques and improved knowledge of the genetics of domestication traits (Zsogon et al., 2022). Demonstrations of this strategy have included the de novo domestication of ground cherry (Physalis pruinose; Lemmon et al., 2018) and development of an allotetraploid rice cultivar from the wild relative Oryza alta (Yu et al., 2021).
4.6. Understudied Indigenous Crops
Human survival relies heavily on a small number of plant species; an estimated 70% of human caloric intake comes from just 15 crops (Zsogon et al., 2022). Understudied Indigenous crops (also known as orphan or underutilized crops) is a term applied to crops that have been overlooked in research efforts and commercial networks, though they may be important food sources regionally, especially in smallholder production (Dwyer et al., 2022). Examples include cowpea (Vigna unguiculata), finger millet (Eleusine coracana subsp. coracana), grain amaranth (Amaranthus hypochondriacus), grass pea (Lathyrus sativus), and African eggplant (Solanum aethiopicum) (Kamenya et al., 2021; Munoz-Amatriain et al., 2017). Because they are often grown in marginal environments, these crops are natural repositories of genetic diversity for stress tolerance. For example, finger millet is reported to possess high levels of resilience to high temperatures and low soil moisture, as well as diversity for biotic stress tolerance and nutritional attributes (Gupta et al., 2017). Among the constraints to increase use of understudied crops, Kamenya et al. (2021) cites the lack of adequate germplasm conservation. Those authors call for coordinated research efforts for understudied crops that take advantage of modern breeding methods such as genomic selection, genome editing, and high throughput phenotyping. In addition to improvement of understudied Indigenous crops, research on these species might also reveal genes that can be integrated into major crop species.
Video 3. “Cowpea diversity: Origins and importance” is a video produced by Colorado State University Center for Science Communication. Cowpeas offer great potential as a crop that can thrive under changing climate conditions.
Video 4. “Cowpea diversity: Producer and consumer traits” is a video produced by Colorado State University Center for Science Communication. Cowpea plants produce protein-rich seeds under stressful environmental conditions.
4.7. Allele mining
Genebanks are widely regarded as treasure troves of genetic diversity that can be exploited in crop improvement schemes. One approach for identifying the beneficial variants among all the non-adaptive gene sequences is known as allele mining, defined by Guerra et al. (2022) as “systematic research, identification and characterization of new allelic variants for one or multiple genes in large and diverse collections”. The approach was employed by Guerra et al. (2022) in a collection of 403 barley cultivars, landraces, and wild relative accessions. Their objective was to find useful variants for frost tolerance and vernalization requirement. These traits might become even more relevant as barley growing areas shift to more northern latitudes (or more southern latitudes in the southern hemisphere) due to climate change. Rather than sequencing entire genomes, the researchers used exome capture and sequencing, which targets just the nucleotide sequences of genes, rather than including sequences between genes. The authors looked at single nucleotide polymorphisms (SNPs) as well as structural variations such as copy number variations and inversions of DNA segments. They reported novel deletions in VRN-H1 associated with a reduced vernalization requirement, and variation at HvCBF14 as the major determinant of frost tolerance.
5. Breeding plants for climate change mitigation
Carbon sequestration can help lower the amount of CO2 in the atmosphere, thereby reducing global climate change. The USGS website provides additional information about carbon sequestration. Efforts to develop agricultural crops with deeper, more extensive root systems, seek to improve not only drought tolerance, but also the plants’ ability to sequester greater amount of carbon. Carbon deposition occurs in root biomass per se, as well as through exudation of carbon-rich compounds into the rhizosphere. Crop species and cultivars within species are known to vary in rooting depth, thus indicating the feasibility of breeding for deeper roots, which decay more slowly than shallower roots (Kell, 2011; Lynch, 2022). The chemical composition of roots has an important impact on their longevity in the soil and thus on their ability to stably sequester carbon; suberin and lignin have been identified as two durable root compounds with especially slow degradation rates (Lynch et al., 2021). A further approach to greater carbon sequestration in root systems is the development of perennial crops to replace annual cultivars, as described for rice (Stokstad, 2022), a wheat wild relative (DeHaan et al., 2020), and other grain crops (Soto-Gomez and Perez-Rodriguez, 2022).
Another strategy for mitigating climate change through genetics is to develop plants with the capacity for biological nitrification inhibition (BNI; Subbarao et al., 2017). The manufacture and use of nitrogen fertilizers for crops is responsible for nearly 80% of global emissions of nitrous oxide (N2O), a potent greenhouse gas (Subbarao et al., 2017). This gas is released to the atmosphere through microbial nitrification and denitrification of nitrogen fertilizer. BNI is the process by which nitrification inhibiting phytochemcials are produced in roots of certain plants and secreted into the rhizosphere, thereby suppressing soil nitrification (Subbarao et al., 2017). Wheat root systems have a weak BNI capacity, but the wild relative species Leymus racemosus was found to have a high capacity for BNI. Subbarao et al. (2021) identified the L. racemosus chromosome segment responsible for BNI, transferred that segment to the standard wheat cultivar ‘Chinese Spring’, and subsequently to elite cultivars. The experimental lines possessing the translocated segment had nearly double the BNI capacity of the controls, as well as higher biomass and grain yields due to reduced nitrogen loss and improved nitrogen nutrition. Grain protein levels and breadmaking qualities were not affected. If equally effective in production environments, BNI crops have the potential to lower N2O emissions, reduce nitrogen use in agriculture, and improve nitrogen use efficiency.
6. Conclusions
Plant breeders have incorporated diversity from genebank collections into many crop cultivars, especially for biotic stress resistance traits. To take advantage of the wealth of conserved PGR more fully, access to genotypic and phenotypic data for traits relevant to climate change will be crucial. Improved understanding of the physiology and molecular biology of abiotic stress tolerance traits, including knowledge of the genes involved, will help direct phenotypic evaluation strategies and will allow gene editing and allele mining approaches to be applied. Root architecture traits offer promise for improving abiotic stress tolerance as well as for mitigating climate change through carbon sequestration. Understudied crops, landraces, and crop wild relatives, which have often been overlooked in the past, deserve more attention as reservoirs of diversity for abiotic stress tolerance. Prebreeding will facilitate the use of PGR in applied breeding programs. Regardless of the specific crop, trait, or methods employed, the diversity represented by PGR has an important role to play in all breeding programs. Ensuring continued access to PGR in ex situ collections is critical, especially with changing climate conditions.
7. References
Araus JL, Kefauver SC. 2018. Breeding to adapt agriculture to climate change: Affordable phenotyping solutions. Current Opinion in Plant Biology 45:237-247. DOI: 10.1016/j.pbi.2018.05.003
Berger JD, Hughes S, Snowball R, Redden B, Bennett SJ, Clements JC, Nawar F. 2013. Strengthening the impact of plant genetic resources through collaborative collection, conservation, characterization, and evaluation: a tribute to the legacy or Dr. Clive Francis. Crop Pasture Science 64:300-311. DOI: 10.1071/CP13023
Blossei J, Gaebelein R, Hammann T, Uptmoor R. 2022. Late blight resistance in wild potato species – Resources for future potato (Solanum tuberosum) breeding. Plant Breeding 141:314-331. DOI: 10.1111/pbr.13023
Bowman BC, Chen J, Zhang J, Wheeler J, Wang Y, et al. 2015. Evaluating grain yield in spring wheat with canopy spectral reflectance. Crop Science 55: 1881-1890. DOI: 10.2135/cropsci2014.08.0533
Brown AHD. 1989. The case for core collections. In: Brown, AHD, Frankel OH, Marshall DR, Williams JT (eds.), The use of plant genetic resources. Cambridge Univ. Press, Cambridge, UK. Available from: researchgate.net/publication/279524326_The_case_for_core_collections
Byrne PF, Volk GM, Gardner C, Gore MA, Simon PW, Smith S. 2018. Sustaining the future of plant breeding: The critical role of the USDA-ARS National Plant Germplasm System. Crop Science 58:451-468. DOI: 10.2135/cropsci2017.05.0303
Crop Genebank Knowledge Base. 2022. Pre-breeding for effective use of plant genetic resources: E-learning course. Accessed on 13 Sep 2022. Available from: cropgenebank.sgrp.cgiar.org/index.php/learning-space-mainmenu-454/training-modules-mainmenu-455/pre-breeding-for-effective-use-of-plant-genetic-resources
Crop Trust. 2022. Crop wild relatives. Crop Trust. Bonn, Germany. Accessed on 13 Sep 2022. Available from: croptrust.org/our-work/supporting-crop-conservation/crop-wild-relatives
DeHaan L, Larson S, Lopez-Marques RL, Wenkel S, Gao C, Palmgren M. 2020. Roadmap for accelerated domestication of an emerging perennial grain crop. Trends in Plant Science 25:525-537. DOI: 10.1016/j.tplants.2020.02.004
Deutsch CA, Tewksbury JJ, Tigchelaar M, Battisti DS, Merrill SC, Huey RB, Naylor RL. 2018. Increase in crop losses to insect pests in a warming climate. Science 361:916-919. DOI: 10.1126/science.aat3466
Díaz-Álvarez EA, Martínez-Zavaleta JP, López-Santiz EE, de la Barrera E, Larsen J, et al. 2021. Climate change can trigger fall armyworm outbreaks: a developmental response experiment with two Mexican maize landraces. International Journal of Pest Management. DOI: 10.1080/09670874.2020.1869347
Dwyer W, Ibe CN, Rhee SY. 2022. Renaming indigenous crops and addressing colonial bias in scientific language. Trends in Plant Science 27:1189-1192. DOI: 10.1016/j.tplants.2022.08.022
Faye JM, Akata EA, Sine B, Diatta C, Cisse N, et al. 2021. Quantitative and population genomics suggest a broad role of stay-green loci in the drought adaptation of sorghum. Plant Genome 15:e20176. DOI: 10.1002/tpg2.20176
FAO. 2015. Coping with climate change – The roles of genetic resources for food and agriculture. Food and Agriculture Organization of the United Nations, Rome. Available from: fao.org/documents/card/es/c/0099d145-f240-4e61-b30e-3d210972ceb8
Fatima T, Arora NK. 2019. Plant growth-promoting rhizospheric microbes for remediation of saline soils. In: Arora NK, Kumar N (eds.) Phyto and rhizo remediation. Springer, Singapore. DOI: 10.1007/978-981-32-9664-0_5
Grogan SM, Anderson J, Baenziger PS, Frels K, Guttieri MJ, et al. 2016. Phenotypic plasticity of winter wheat heading date and grain yield across the U.S. Great Plains. Crop Science 56:2223-2236. DOI: 10.2135/cropsci2015.06.0357
Guerra D, Morcia C, Badeck F, Rizza F, Delbono S, et al. 2022. Extensive allele mining discovers novel genetic diversity in the loci controlling frost tolerance in barley. Theoretical and Applied Genetics 135:553-569. DOI: 10.1007/s00122-021-03985-x
Gupta SM, Arora S, Mirza N, Pande A, Lata C, et al. 2017. Finger millet: A “certain” crop for an “uncertain” future and a solution to food insecurity and hidden hunger under stressful environments. Frontiers in Plant Science 8:643. DOI: 10.3389/fpls.2017.00643
Huang LL, Xue FS, Chen C, Guo X, Tang JJ, et al. 2021. Effects of temperature on life history traits of the newly invasive fall armyworm, Spodoptera frugiperda in Southeast China. Ecology and Evolution 11:5255-5264. DOI: 10.1002/ece3.7413
Irish BM, Volk GM. 2023. Climate change affects plant interactions with pollinators, pathogens, and pests. In: Volk GM, Moreau TL, Byrne PF. Conserving and Using Climate-Ready Plant Collections. Fort Collins, Colorado: Colorado State University. Available from: colostate.pressbooks.pub/climatereadyplantcollections/chapter/pollinators-pathogens-and-pests
Kamenya SN, Mikwa EO, Song B, Odeny DA. 2021. Genetics and breeding for climate change in orphan crops. Theoretical and Applied Genetics 134:1787-1815. DOI: 10.1007/s00122-020-03755-1
Karavolias NG, Horner W, Abugu MN, Evanega SN. 2021. Application of gene editing for climate change in agriculture. Frontiers in Sustainable Food Systems 5:685801. DOI: 10.3389/fsufs.2021.685801
Karki HS, Jansky SH, Halterman DA. 2021. Screening of wild potatoes identifies new sources of late blight resistance. Plant Disease 2:368-376. DOI: 10.1094/PDIS-06-20-1367-RE
Kaur N, Alok A, Shivani, Kumar P, Kaur N, et al. 2020. CRISPR/Cas9 directed editing of lycopene epsilon-cyclase modulates metabolic flux for b-carotene biosynthesis in banana fruit. Metabolic Engineering 59:76-86. DOI: 10.1016/j.ymben.2020.01.008
Kell DB. 2011. Breeding crop plants with deep roots: their role in sustainable carbon, nutrient and water sequestration. Annals of Botany 108:407-418. DOI: 10.1093/aob/mcr175
Kim YC, Glick BR, Bashan Y, Ryu CM. 2012. Enhancement of plant drought tolerance by microbes. In: Aroca R (ed.) Plant responses to drought stress. Springer, Heidelberg. DOI: 10.1007/978-3-642-32653-0_15
Kumar N, Galli M, Ordon J, Stuttmann J, Kogel K, et al. 2018. Further analysis of barley MORC1 using a highly efficient RNA-guided Cas9 gene-editing system. Plant Biotechnology Journal 16:1892-1903. DOI: 10.1111/pbi.12924
Kunert K, Vorster BJ. 2020. In search for drought-tolerant soybean: Is the slow-wilting phenotype more than just a curiosity? Journal of Experimental Botany 71:457-460. DOI: 10.1093/jxb/erz235
Leigh FJ, Wright TIC, Horsnell RA, Dyer S, Bentley AR. 2022. Progenitor species hold untapped diversity for potential climate-responsive traits for use in wheat breeding and crop improvement. Heredity 128:291-303. DOI: 10.1038/s41437-022-00527-z
Lemmon ZH, Reem NT, Dalrymple J, Soyk S, Swartwood KE, et al. 2018. Rapid improvement of domestication traits in an orphan crop by genome editing. Nature Plants 4:766-770. DOI: 10.1038/s41477-018-0259-x
Lynch JP. 2022. Harnessing root architecture to address global challenges. The Plant Journal 109:415-431. DOI: 10.1111/tpj.15560
Lynch JP, Strock CF, Schneider HM, Singh Sidh J, Ajmera I, Galindo-Castañeda T, Klein SP, Hanlon MT. 2021. Root anatomy and soil resource capture. Plant Soil 466:21-63. DOI: 10.3929/ethz-b-000497409
Mackay MC, Street KA, Mitrofanova O, Konopka J, Berger JD. 2007. The objective exploitation of plant genetic resources using the focused identification of germplasm strategy. In: ‘2nd International Vavilov Conference: Crop Genetic Resources in the 21st Century: Current Status, Problems and Prospects’. p. 26-30.
Martini JWR, Molnar TL, Crossa J, Hearne SJ. Pixley KV. 2021. Opportunities and challenges of predictive approaches for harnessing the potential of genetic resources. Frontiers in Plant Science 12:674036. DOI: 10.3389/fpls.2021.674036
Merchuk-Ovnat L, Barak V, Fahima T, Ordon F, Lidzbarsky GA, et al. 2016. Ancestral QTL alleles from wild emmer wheat improve drought resistance and productivity in modern wheat cultivars. Frontiers in Plant Science 7:452. DOI: 10.3389/fpls.2016.00452
Millet E, Steffenson BJ, Prins R, Sela H, Przewieslik-Allen AM, et al. 2017. Genome targeted introgression of resistance to African stem rust from Aegilops sharonensis into bread wheat. The Plant Genome 10:3. DOI: 10.3835/plantgenome2017.07.0061
Munoz-Amatriain M, Mirebrahim H, Xu P, Wanamaker SI, Luo MC, et al. 2017. Genome resources for climate-resilient cowpea, an essential crop for food security. The Plant Journal 89:1042-1054. DOI: 10.1111/tpj.13404
Ober ES, Alahmad S, Cockram J, Forestan C, Hickey LT, et al. 2021. Wheat root systems as a breeding target for climate resilience. Theoretical and Applied Genetics 134:1645-1662. DOI: 10.1007/s00122-021-03819-w
Pixley KV, Falck-Zepeda JB, Paarlberg RL, Phillips PWB, Slamet-Loedin IH, et al. 2022. Genome-edited crops for improved food security of smallholder farmers. Nature Genetics 54:364-367. DOI: 10.1038/s41588-022-01046-7
Placido DF, Sandhu J, Sato SJ, Nersesian N, Quach T, et al. (2020) The LATERAL ROOT DENSITY gene regulates root growth during water stress in wheat. Plant Biotechnology Journal 18:1955-1968. DOI: 10.1111/pbi.13355
Prank M, Kenaley SC, Bergstrom GC, Acevedo M, Mahowald NM. 2019. Climate change impacts the spread potential of wheat stem rust, a significant crop disease. Environmental Research Letters 14:124053. DOI: 10.1088/1748-9326/ab57de
Schmidt JE, Bowles TM, Gaudin ACM. 2016. Using ancient traits to convert soil health into crop yield: Impact of selection on maize root and rhizosphere function. Frontiers in Plant Science 7:373. DOI: 10.3389/fpls.2016.00373
Schaffasz A, Windpassinger S, Friedt W, Snowdon R, Wittkop B. 2019. Sorghum as a novel crop for Central Europe: Using a broad diversity set to dissect temperate-adaptation. Agronomy 9:535. DOI: 10.3390/agronomy9090535
Shi J, Gao H, Wang H, Lafitte HR, Archibald RL, et al. 2017. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnology Journal 15:207-216. DOI: 10.1111/pbi.12603
Sinclair TR, Messina CD, Beatty A, Samples M. 2010. Assessment across the United States of the benefits of altered soybean drought traits. Agronomy Journal 102:475-482. DOI: 10.2134/agronj2009.0195
Singh GM, Xu J, Schaefer D, Day R, Wang Z, et al. 2021. Maize diversity for fall armyworm resistance in a warming world. Crop Science 62:1-19. DOI: 10.1002/csc2.20649
Smith WR, Mengistu A, Nelson RL, Paris RL. 2008. Identification of soybean accessions with high germinability in high-temperature environments. Crop Science 48:2279-2288. DOI: 10.2135/cropsci2008.01.0026
Soto-Gomez D, Perez-Rodriguez P. 2022. Sustainable agriculture through perennial grains: Wheat, rice, maize, and other species. A review. Agriculture, Ecosystems & Environment 325:107747. DOI: 10.1016/j.agee.2021.107747
Stokstad E. 2022. Perennial rice could be a ‘game changer’. Science 378:586. DOI: 10.1126/science.adf7191
Subbarao G, Arango J, Masahiro K, Hooper AM, Yoshihashi T, et al. 2017. Genetic mitigation strategies to tackle agricultural GHG emissions: The case for biological nitrification inhibition technology. Plant Science 262:165-168. DOI: 10.1016/j.plantsci.2017.05.004
Subbarao GV, Kishii M, Bozal-Leorric A, Ortiz-Monasterio I, Gao X, et al. 2021. Enlisting wild grass genes to combat nitrification in wheat farming: A nature-based solution. Proceedings of the National Academy of Science 118:e2106595118. DOI: 10.1073/pnas.2106595118
Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, et al. 2013. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nature Genetics 45:1097-1102. DOI: 10.1038/ng.2725
Ullstrup AJ. 1972. The impacts of the Southern Corn Leaf Blight epidemics of 1970–1971. Annual Review of Phytopathology 10:37-50. DOI: 10.1146/annurev.py.10.090172.000345
Vikram P, Franco J, Burgueño J, Li H, Seghal D, et al. 2021. Strategic use of Iranian bread wheat landrace accessions for genetic improvement: Core set formulation and validation. Plant Breeding 140:87-99. DOI: 10.1111/pbr.12885
Volk, GM, Byrne PF, Coyne CJ, Flint-Garcia S, Reeves PA, et al. 2021. Integrating genomic and phenomic approaches to support plant genetic resources conservation and use. Plants 10:2260. DOI: 10.3390/plants10112260
Wang W, Pan Q, He F, Akhunova A, Chao S, et al. 2018. Transgenerational CRISPR-Cas9 activity facilitates multiplex gene editing in allopolyploid wheat. The CRISPR Journal 1:65-74. DOI: 10.1089/crispr.2017.0010
Ye H, Song L, Schapaugh WT, Ali ML, Sinclair TR, et al. 2020. The importance of slow canopy wilting in drought tolerance in soybean. Journal of Experimental Botany 71:642-652 DOI: 10.1093/jxb/erz150
Yu G, Matny O, Champouret N, Steuernagel B, Moscou MJ, et al. 2022. Aegilops sharonensis genome-assisted identification of stem rust resistance gene Sr62. Nature Communications 13:1607. DOI: 10.1038/s41467-022-29132-8
Yu H, Lin T, Meng X, Du H, Zhang J, et al. 2021 A route to de novo domestication of wild allotetraploid rice. Cell, 184:1156-1170.e14. DOI: 10.1016/j.cell.2021.01.013
Zhang A, Liu Y, Wang F, Li T, Chen Z, et al. 2019. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 39:47. DOI: 10.1007/s11032-019-0954-y
Zhang H, Zhang J, Lang Z, Botella JR, Zhu JK. 2017. Genome editing—principles and applications for functional genomics research and crop improvement. Critical Reviews in Plant Sciences 36:291-309. DOI: 10.1080/07352689.2017.1402989
Zsogon A, Peres LEP, Xiao Y, Yan J, Fernie AR. 2022. Enhancing crop diversity for food security in the face of climate uncertainty. The Plant Journal 109:402-414. DOI: 10.1111/tpj.15626
8. acknowledgments
Citation: Byrne PF. 2023. Plant breeding for climate change: Opportunities for adaptation and mitigation. In: Volk GM, Moreau TL, Byrne PF. Conserving and Using Climate-Ready Plant Collections. Fort Collins, Colorado: Colorado State University. Available from: https://colostate.pressbooks.pub/climatereadyplantcollections/chapter/plant-breeding-for-climate-change/
This eBook chapter was developed through a collaboration among USDA-Agricultural Research Service, Colorado State University, and the University of British Columbia Botanic Garden, with additional funding from the USDA-NIFA-Higher Education Challenge Grant (2020-70003-303930).
Editors: Katheryn Chen, Gayle M. Volk
Videographer: CSU Center for Science Communication
a crop variety produced by scientific breeding or farmer selection methods
in a climate change context, taking action to prepare for and adjust to both the current and projected impacts of climate change
in a climate change context, actions that reduce or prevent greenhouse gas emissions or that lower the amount of greenhouse gases in the atmosphere
the organs and tissues (seeds, fruits, cuttings, pollen, tissue cultures, etc.) by which plants can be propagated (synonymous with plant germplasm)
genetically distinct uniquely identifiable samples of plant genetic resources within a genebank
observable characteristic of an organism
allelic constitution of an organism at one or more loci
basic information about the origin of a genebank accession, such as collecting site or pedigree
introgression of desired genes from wild plant species (or crop wild relatives) into breeding lines; enrichment of a breeding population for particular genes and/or traits; and development of novel breeding populations
the organs and tissues (seeds, fruits, cuttings, pollen, tissue cultures, etc.) by which plants can be propagated (synonymous with plant genetic resources)
a local plant variety, usually genetically heterogeneous, with distinctive characteristics arising from selection over time for adaptation to localized conditions or food preferences
the wild undomesticated plants most closely related to agricultural crops that potentially possess useful traits/genes/alleles for crop improvement
transfer of genetic information from one taxon (or parent) to another through hybridization and repeated backcrossing to one of the taxa (or parents)
Latin term meaning “from the beginning”
necessary chilling prior to growth of dormant or quiescent plant materials (such as for seed germination)
the capture and storage of atmospheric carbon dioxide
conservation of plant genetic resources outside their natural habitat

