From Wild Species to Landraces and Cultivars
Patrick Byrne, Dept. of Soil and Crop Sciences, Colorado State University, Fort Collins, Colorado 80523 (Patrick.Byrne@colostate.edu)
Christopher Richards, USDA-ARS National Laboratory for Genetic Resources Preservation, 1111 S. Mason St., Fort Collins, Colorado, 80521
Gayle M. Volk, USDA-ARS National Laboratory for Genetic Resources Preservation, 1111 S. Mason St., Fort Collins, Colorado, 80521
Outline
1. Introduction
The wild relatives of domesticated plant species usually do not possess all the desirable traits for routine agricultural production and use. This chapter offers an overview of the process of early crop domestication from thousands of years ago through to the progress in plant improvement by modern-day plant breeders. An understanding of crop domestication and plant breeding provides a context for the value, importance, and use of crop wild relatives that are conserved either in situ or in genebanks.
2. Early crop domestication
One of the landmarks of human history is the transition from nomadic hunter-gatherer societies to settled agriculture-based societies, the so-called neolithic revolution. A key component of this transition was the domestication of wild species into cultivated crops capable of supporting higher population densities (Harlan, 1992; Schwanitz, 1966). Crop domestication from wild species began about 8000 to 10,000 years ago independently in many parts of the world. Some of the more notable centers of domestication were the Fertile Crescent of the Middle East (wheat, barley, lentil, and chickpea), Mesoamerica (maize or corn, chiles, squash, and common bean), the Andean region (potato, tomato, and a second center of origin for common bean), and Southeast Asia (rice, millet, and soybean).
Early farmers selected for traits either deliberately or unintentionally that made the wild plants more suitable for human needs (Figure 1). These included characteristics that improved yield (more or bigger harvested product; reduced branching), made agricultural production easier (loss of seed dormancy; retention of seed on the plant), or improved product quality (reduced bitterness or toxic properties; easier processing). The term ‘domestication syndrome’ has been used to encompass the collection of traits that were commonly selected (Gepts, 2018).
 Figure 1. Comparison of seeds of wild progenitor species (left) and their domesticated counterparts (right). Note change in seed size, as well as reduced pigmentation in some cases. From top to bottom: pistachio, coffee, soybean, barley, North American wild rice (Zizania palustris), and sorghum. Photo credit: Christina Walters.
Figure 1. Comparison of seeds of wild progenitor species (left) and their domesticated counterparts (right). Note change in seed size, as well as reduced pigmentation in some cases. From top to bottom: pistachio, coffee, soybean, barley, North American wild rice (Zizania palustris), and sorghum. Photo credit: Christina Walters.
Many of the domestication traits are characterized by simple inheritance, i.e., they are controlled by one or two genes with large effects rather than by multiple genes. The simple inheritance of domestication traits would have facilitated their selection and incorporation into wild populations (Gepts, 2018). Domestication most likely occurred over an extended period of time, with ongoing cross-pollination between wild and domesticated populations. Because the selected plants represented just a small fraction of the total wild population, domestication was often accompanied by a reduction of genetic diversity (see Figure 3). Some of the traits involved in the transition from wild teosinte to domesticated maize are shown in Video 1, “Corn Domestication”.
Video 1. Corn domestication.
As early farmers migrated into new territories, they brought their seeds, tubers, and rootstocks with them. The crops would have encountered a variety of soil types, weather patterns, insect pests, and pathogens. In response to the new environments, farmers would have selected for plants that were better adapted to the local conditions. Thus arose ‘landraces’, traditional varieties selected by farmers for adaptation to local conditions and food preferences. These varieties typically have low yield potential, but are often genetically diverse and stress-resilient. Many so-called ‘heirloom varieties’ are examples of landraces. Typically landraces are genetically heterogeneous (non-uniform), and are therefore an important source of genetic diversity for crop improvement. Landraces are usually less diverse than their wild ancestors, but more diverse than cultivars produced during the modern era of plant breeding. This is shown conceptually in Figure 2, and on the basis of DNA analysis in common bean (Figure 3).
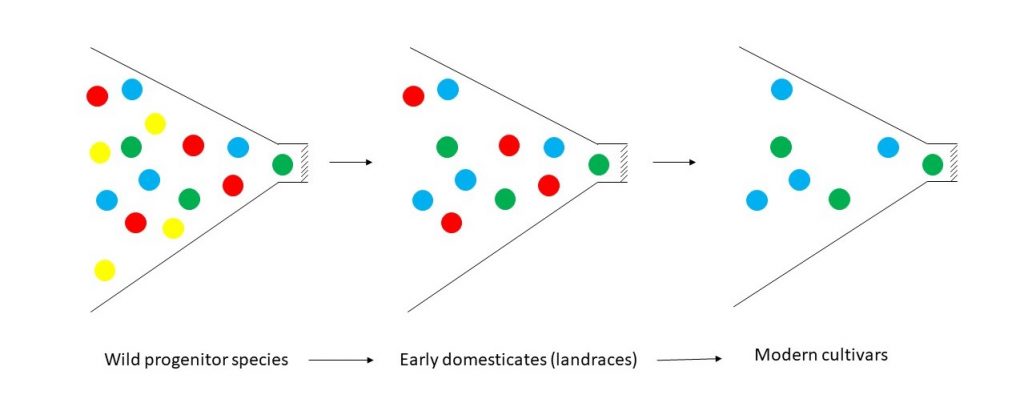 Figure 2. Conceptual representation of a reduction in genetic diversity (the bottleneck effect) as wild species were domesticated, and a further reduction that occurred with development of modern cultivars. Colors represent the diversity of alleles within a population. Redrawn from Tanksley and McCouch (1997). Rather than abrupt changes as indicated by the pronounced bottlenecks here, domestication and crop improvement are now viewed as leading to a gradual loss of diversity over a long period of time.
Figure 2. Conceptual representation of a reduction in genetic diversity (the bottleneck effect) as wild species were domesticated, and a further reduction that occurred with development of modern cultivars. Colors represent the diversity of alleles within a population. Redrawn from Tanksley and McCouch (1997). Rather than abrupt changes as indicated by the pronounced bottlenecks here, domestication and crop improvement are now viewed as leading to a gradual loss of diversity over a long period of time.
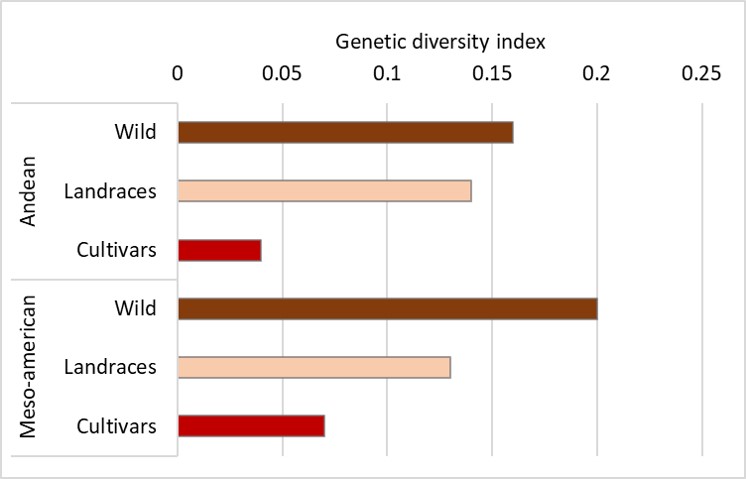 Figure 3. Reduction of genetic diversity during evolution of common bean (Phaseolus vulgaris) in the Andean and Meso-American centers of origin. Redrawn from Gepts (2018).
Figure 3. Reduction of genetic diversity during evolution of common bean (Phaseolus vulgaris) in the Andean and Meso-American centers of origin. Redrawn from Gepts (2018).
Video 2. How is the genetic diversity of a species affected by domestication and breeding?
3. A brief history of plant breeding
Plant breeding has been defined as the “art and science of improving the heredity of plants for the benefit of humankind” (Sleper and Poehlman, 2006). Plant breeders engage in a cyclical process of collecting or generating heritable variation for traits of interest, evaluating that variation, and selecting superior individuals. Empirical or intuitive plant breeding was practiced for many centuries by observant farmers who recognized the value of particular traits and selected a portion of their harvest to plant the following season (Murphy, 2007). Although they did not understand the scientific basis of trait inheritance, they were able to effect crop improvement through their selections.
Scientific plant breeding dates to around 1900 when Gregor Mendel’s work on trait inheritance was rediscovered. Breeders applied Mendel’s principles to the selection of qualitative traits (those controlled by one or two major genes), and developed increasingly sophisticated methods to improve quantitative traits (those controlled by many genes, each having a small effect). The concept of heritability (the proportion of total variability due to genetics as opposed to environmental factors) was elucidated and helped guide evaluation and selection strategies. Field plot design and statistical analysis methods further improved the accuracy with which traits were evaluated and candidate varieties were compared. The observation of hybrid vigor and its converse, inbreeding depression, led to development of the hybrid seed industry, initially in maize (Video 3), and later in other crops.
Video 3. Hybrid vigor in corn.
Beginning in the 1960’s, the Green Revolution transformed wheat and rice production through the incorporation of semi-dwarfing genes into taller varieties. Reduced plant height meant that more photosynthate could be directed to grain production rather than stems and leaves. Shorter, stiffer stalks also reduced the amount of lodging (bending over of the stems). Although the Green Revolution averted famine and saved many lives, it is not without controversy, as described in the IFPRI brief (2002).
Another revolution in plant breeding occurred with the development and exploitation of DNA molecular markers beginning in the 1980’s (Jain and Brar, 2010). Marker technologies permitted determination of genetic relationships (e.g., between crops and their wild progenitors); fingerprinting of cultivars and breeding lines; elucidating the biochemical basis of trait expression; locating and characterizing quantitative trait loci (QTL, genes with small effects that contribute to quantitative trait inheritance); and marker-assisted selection based on DNA patterns rather than trait observation. Individual markers can be useful if they are closely linked to a major gene. Many breeding programs are also now applying genomic selection, whereby the additive effects of thousands of markers are incorporated into selection models (Voss-Fels et al., 2019).
Although genomic technologies have enabled high-throughput genotyping of plants’ DNA patterns, phenotyping (trait evaluation) of large numbers of breeding lines remains a bottleneck. A number of technologies have been developed to relieve this constraint, including the deployment of hand-held, tractor mounted, and aerial sensors of crop canopy spectral reflectance and temperature (Araus and Cairns, 2014; Zhao et al., 2019) (Figure 4).
 Figure 4. (Left) A high-clearance tractor with multiple attached sensors is used to phenotype wheat in Dr. Jesse Poland’s program at Kansas State University. Photo credit: Pat Byrne. (Right) A quad-copter drone (or UAV, unmanned aerial vehicle) captures spectral reflectance data in a Colorado State University wheat field trial. Photo credit: Scott Reid.
Figure 4. (Left) A high-clearance tractor with multiple attached sensors is used to phenotype wheat in Dr. Jesse Poland’s program at Kansas State University. Photo credit: Pat Byrne. (Right) A quad-copter drone (or UAV, unmanned aerial vehicle) captures spectral reflectance data in a Colorado State University wheat field trial. Photo credit: Scott Reid.
Another landmark in the history of cultivar development was the commercialization of transgenic (genetically engineered or genetically modified) crops beginning in the mid-1990’s. These are crops that contain genes transferred from other species by recombinant DNA technology using laboratory protocols. Transgenic crops have had a major impact in a relatively small number of crops, notably corn or maize, soybean, cotton, canola, alfalfa, and sugarbeet. Most transgenic cultivars currently on the market have been improved for herbicide tolerance or insect resistance, though virus resistant, non-browning, and drought tolerant cultivars have also been commercialized. The U.S. National Academy of Sciences, Engineering, and Medicine (2016) published a comprehensive report on the impact of transgenic crops, including on health and environmental safety. With regard to crop wild relatives, a concern has been the potential transfer of transgenes to wild populations. This is a risk that will vary with each crop and geographic region, and needs to be carefully considered before approval of a transgenic crop for a new region.
The genetic diversity conserved in the world’s genebanks, especially for wild relatives of crops, is a largely untapped resource for crop improvement (Byrne et al., 2018). Challenges to the greater use of crop wild relatives in cultivar development were described by Dempewolf et al. (2017), based on consultations with experts in 24 crop communities. Those challenges include insufficient phenotypic and genotypic data on crop wild relative accessions; ploidy differences and other hybridization barriers between wild species and cultivated germplasm; the existence of sufficient variation in cultivated germplasm of some crops; inferiority of crop wild relatives for desired traits; linkage drag; and inadequate human or financial resources to conduct the necessary research and development. Creative application of technology and appropriate policy changes will help overcome these challenges to unlock the diversity of genebank collections for crop improvement. Guidelines for sampling in the wild, evaluating collections for favorable variation, and incorporating the beneficial genetics into breeding programs are provided by Hübner and Kantar (2021). Kashyap et al. (2022) provide examples of successful use of wild relatives in crop improvement and discuss strategies to facilitate introgression of wild germplasm into breeding programs.
4. References
Araus JL, Cairns JE. 2014. Field high-throughput phenotyping: The new crop breeding frontier. Trends in Plant Science 19:52-61.
Byrne PF, Volk GM, Gardner C, Gore MA, Simon PW, Smith S. 2018. Sustaining the future of plant breeding: The critical role of the USDA-ARS National Plant Germplasm System. Crop Science 58:451-468.
Dempewolf H, Baute G, Anderson J, Kilian B, Smith C, Guarino L. 2017. Past and future use of wild relatives in crop breeding. Crop Science 57:1070-1082.
Gepts P. 2018. The domestication of our food crops. In: Chrispeels MJ, Gepts P, (editors). Plants, Genes & Agriculture: Sustainability through Biotechnology. Oxford University Press, Cary, North Carolina.
Harlan JR. 1992. Crops & Man, 2nd edition. American Society of Agronomy and Crop Science Society of America, Madison, WI.
Hübner S, Kantar MB. 2021. Tapping diversity from the wild: From sampling to implementation. Frontiers in Plant Science 12:626565. https://doi.org/10.3389/fpls.2021.626565
IFPRI. 2002. Green Revolution: Curse or blessing? International Food Policy Research Institute, Washington, D.C.
Jain SM, Brar DS (editors). 2010. Molecular Techniques in Crop Improvement, 2nd edition. Springer, Dordrecht.
Kashyap A, Garg P, Tanwar K, Sharma J, Gupta NC, Ha PTT, Bhattacharya RC, Mason AS, Rao M. 2022. Strategies for utilization of crop wild relatives in plant breeding programs. Theoretical and Applied Genetics 135:4151-4167. https://doi.org/10.1007/s00122-022-04220-x
Murphy DJ. 2007. Plant Breeding and Biotechnology: Societal Context and the Future of Agriculture. Cambridge University Press, Cambridge, UK.
National Academies of Sciences, Engineering, and Medicine. 2016. Genetically Engineered Crops: Experiences and Prospects. The National Academies Press, Washington, D.C. https://doi.org/10.17226/23395
Schwanitz F. 1966. The Origin of Cultivated Plants. Harvard University Press, Cambridge, MA.
Sleper DA, Poehlman JM. 2006. Breeding Field Crops, 5th edition. Blackwell Publishing, Ames, IA.
Tanksley SD, McCouch SR. 1997. Seed banks and molecular maps: unlocking genetic potential from the wild. Science 277:1063-1066.
Voss-Fells KP, Cooper M, Hayes B. 2019. Accelerating crop genetic gains with genomic selection. Theoretical and Applied Genetics 132:669-686.
Zhao CJ, Zhang Y, Du JJ, Guo XY, Wen WL, Gu SH, Wang JL, Fan JC. 2019. Crop phenomics: Current status and perspectives. Frontiers in Plant Science 10:714.
5. Acknowledgments
Citation: Byrne P, Richards C, Volk GM. 2020. From Wild Species to Landraces and Cultivars. In: Volk GM, Byrne P (Eds.) Crop Wild Relatives and their Use in Plant Breeding. Fort Collins, Colorado: Colorado State University. Date accessed. Available from https://colostate.pressbooks.pub/cropwildrelatives/chapter/from-wild-species-to-landraces-and-cultivars/
This training module was made possible in part by funding from USDA-ARS, Colorado State University, IICA-PROCINORTE (procinorte.net), and the United States Agency for International Development (USAID).
Chapter editors: Emma Balunek, Katheryn Chen, Gayle Volk
Videographer: Mike May
This project was funded in part by the National Academy of Sciences (NAS) and USAID, and any opinions, findings, conclusions, or recommendations expressed in such are those of the authors alone, and do not necessarily reflect the views of USAID or NAS.


