Case Study: Wheat Domestication and Breeding
Patrick Byrne, Dept. of Soil and Crop Sciences, Colorado State University, Fort Collins, Colorado 80523 (Patrick.Byrne@colostate.edu)
Outline
- Introduction
- A and B genome ancestors and A x B hybridization
- D genome ancestor and AB x D hybridization
- Synthetic hexaploid wheat
- Useful genes from wild wheat relatives
- References
- Acknowledgments
1. Introduction
The domestication of wheat (Triticum aestivum L.) from wild grasses in the Middle East is a fascinating story that resulted in one of the world’s most important and widespread crops. It is estimated that wheat provides about 20% of the energy in global diets and about the same percentage of protein (WHEAT, 2014). Wheat is primarily a cool season crop, but is broadly adapted to many types of growing conditions, both irrigated and rainfed, and is especially important as a staple crop in semi-arid conditions. Part of wheat’s adaptability is due to the fact that it occurs both as winter habit varieties (requiring a cold period of 6 to 8 weeks to trigger reproduction), and spring habit varieties (not requiring the cold period). Wheat geneticists and breeders have a long history of identifying useful genes in wild wheat relatives and incorporating them into improved varieties.
2. A and B genome ancestors and A x B hybridization
Common wheat is a hexaploid species with three sets of similar but distinct chromosomes. These chromosome sets are designated the A, B, and D genomes, each with seven pairs of chromosomes. Thus, hexaploid wheat contains 3 genomes x 7 pairs = 21 pairs or 42 chromosome total. Each genome originated in a different annual diploid grass species in the Fertile Crescent of the Middle East (Figure 1).
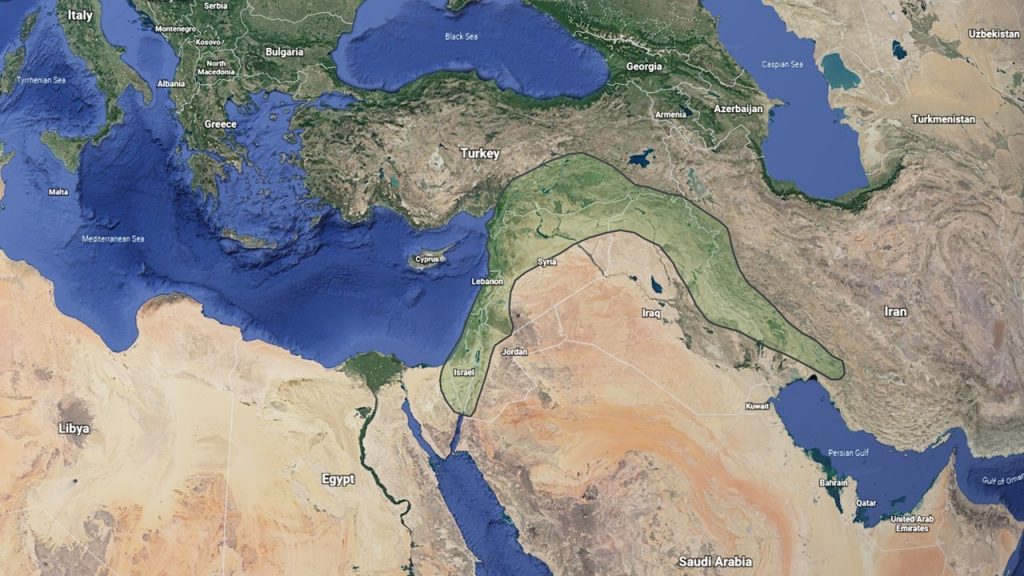 Figure 1. The Fertile Crescent of the Middle East, where wheat and many other crops were domesticated.
Figure 1. The Fertile Crescent of the Middle East, where wheat and many other crops were domesticated.
 Figure 2. The combination of the A, B, and D genomes led to common bread wheat, which has all three genomes. From left to right, A genome, Triticum urartu; B genome, Aegilops speltoides ligustica; D genome, Aegilops tauschii; and A+B+D genome, Triticum aestivum. Photo credit: Pat Byrne.
Figure 2. The combination of the A, B, and D genomes led to common bread wheat, which has all three genomes. From left to right, A genome, Triticum urartu; B genome, Aegilops speltoides ligustica; D genome, Aegilops tauschii; and A+B+D genome, Triticum aestivum. Photo credit: Pat Byrne.
The A genome ancestor of wheat is Triticum urartu, and the B genome is thought to have originated with a close relative of Aegilops speltoides. Hybridization between these progenitors less than one million years ago (Marcussen et al., 2014) gave rise to the tetraploid species Triticum turgidum ssp. dicoccoides, known as wild emmer, having the AABB genome constitution. This wild species was domesticated to form emmer wheat (Triticum turgidum ssp. dicoccum), which gave rise to durum or pasta wheat (Triticum turgidum ssp. durum). Both emmer and durum wheats are tetraploids with genome designation AABB.
A separate lineage of the A genome led to domesticated einkorn wheat (Triticum monococcum ssp. monococcum), which is still grown in remote parts of Turkey, Italy, and Spain. The genome of this diploid species is usually designated AmAm to distinguish it from the lineage that led to hexaploid wheat.
Video 1. Dr. Patrick Byrne shows examples of the A, B, and D genome ancestors of wheat, describing wheat’s evolutionary history, and explaining how wild wheats are relevant to modern wheat breeding.
Video 2. Dr. Patrick Byrne highlights examples of contrasting traits in modern wheat and its wild ancestors or heirloom varieties, providing insight into the history of wheat breeding. The featured traits are plant height, harvest index, lodging, and shattering.
Video 3. Dr. Patrick Byrne shows harvested heads of the A, B, and D ancestors of wheat, and explains the strategy of creating synthetic hexaploid wheat to increase diversity of cultivated wheat.
3. D genome ancestor and AB x D hybridization
Wheat’s D genome ancestor is the goatgrass Aegilops tauschii, which occurs across a broad range of the Middle East and Asia from Syria to China. DNA analysis of wheat and its wild relatives led Marcussen et al. (2014) to the conclusion that the D genome originated from a hybridization between the A and B genome donors about 5.5 million years ago. Much later, another hybridization occurred between domesticated emmer wheat (AABB) and Ae. tauschii (DD), giving rise to hexaploid common or bread wheat (T. aestivum), with the AABBDD genome designation. This hybridization is believed to have occurred only 8000 to 10,000 years ago, most likely in an area south of the Caspian Sea in present-day Iran (Gepts, 2018). The D genome provided wheat with adaptability to the harsh environmental conditions of Central Asia, as well as proteins that improved the breadmaking properties of the flour.
The hybridization that led to common wheat is believed to have involved a very limited number of Ae. tauschii plants, creating a so-called bottleneck effect where only a limited amount of D-genome variation became incorporated into the hexaploid crop. Thus, the D genome is the least genetically diverse of wheat’s three genomes. The diversity present across the vast range of Ae. tauschii is mostly absent in today’s wheat and until recently has been largely unexplored as a source of useful variation.
Video 4. Dr. Patrick Byrne shows the diverse growth habits of wheat’s D-genome progenitor, Aegilops tauschii, and discusses the processes used to cross Ae. tauschii with bread wheat.
4. Synthetic hexaploid wheat
A creative strategy for addressing the impoverished D genome diversity of common wheat is the development of synthetic hexaploid wheats (McFadden and Sears, 1946; Trethowan and Mujeeb-Kazi, 2008; Ogbonnaya et al., 2013). This strategy is an attempt to recreate the evolutionary history of wheat by intentionally crossing tetraploid (AABB) wheat with Ae. tauschii (DD), but using a much broader range of Ae. tauschii accessions than occurred in nature. Ae. tauschii is an attractive species for introgression into common wheat, because its genome readily pairs and recombines with wheat’s D genome chromosomes. The International Maize and Wheat Improvement Center (CIMMYT) has been a leader in producing and utilizing synthetic hexaploids, with over 1,500 synthetics developed (Mujeeb-Kazi and Hettel, 1995; Rosyara et al., 2019).
The process of developing synthetic hexaploid wheat is diagrammed in Figure 3. Because the initial product of fertilization between the tetraploid and diploid species is inviable after a few weeks, an embryo rescue procedure must be used in many cases. This involves dissecting out the young embryo and growing it in tissue culture to produce a triploid (ABD) plant. The chromosome number of this plant is then doubled by treating with colchicine to produce AABBDD plants, the same as common wheat. Other methods for incorporating wild species diversity into wheat include direct crossing of Ae. tauschii or wild tetraploid accessions to hexaploid wheat, as explained in Ogbonnaya et al. (2013).
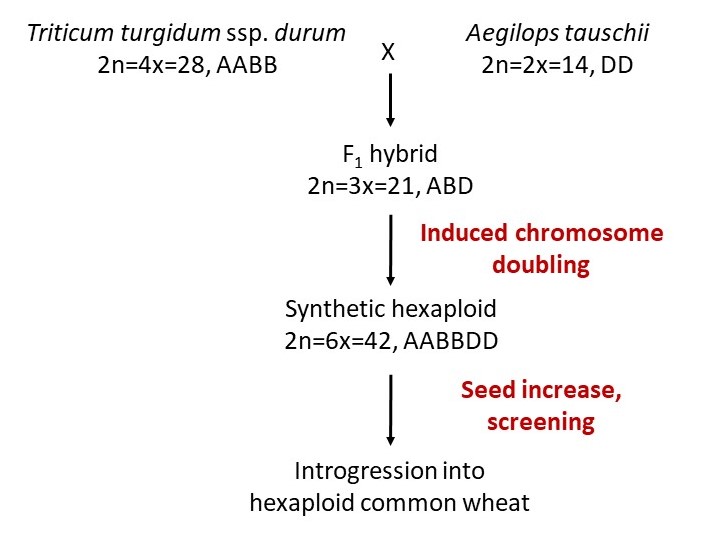 Figure 3. Development of synthetic hexaploid wheat (Ogbonnaya et al., 2013).
Figure 3. Development of synthetic hexaploid wheat (Ogbonnaya et al., 2013).
Synthetic hexaploids are not directly useful for wheat production because they are typically not adapted to the target growing area and they express wild species traits, such as poor threshing (i.e., the seeds do not separate easily from the enclosing glume tissue) and excessive lodging (Figure 3). Therefore, the synthetic lines are typically crossed to an adapted wheat cultivar, then backcrossed once or twice to the adapted cultivar, followed by selection. This results in breeding lines (sometimes called synthetic-derived lines) that have mostly adapted genomes with segments of synthetic hexaploid chromosomes interspersed. These lines can be grown and evaluated like normal wheat in hopes of finding improvements in qualitative or quantitative traits due to the inserted synthetic hexaploid segments.
 Figure 4. Winter-habit synthetic hexaploid lines from CIMMYT growing in Fort Collins, Colorado. Photo credit: Pat Byrne.
Figure 4. Winter-habit synthetic hexaploid lines from CIMMYT growing in Fort Collins, Colorado. Photo credit: Pat Byrne.
CIMMYT has done extensive evaluation of its synthetic-derived lines (Dreccer et al., 2007; Lage and Trethowan, 2008; Rosyara et al., 2019). Ogbonnaya et al., 2013 state that in the 7 years previous to their review, 17% of wheat lines in all of CIMMYT’s international trials were synthetic-derived, but that number increased to 35% for the Semiarid Wheat Yield Trial. In a study of 253 synthetic-derived lines relevant to CIMMYT’s breeding and testing program, the average marker-based estimate of the Ae. tauschii contribution to those lines was 17.4% (Rosyara et al., 2019). Over 80 synthetic-derived lines have been released as cultivars, notably Sokoll and Vorobey (Rosyara et al., 2019).
Synthetic hexaploid lines have been shown to be useful sources of variation for abiotic and biotic stress tolerance, as well as agronomic and novel grain quality traits. Their improved yield performance under drought stress conditions appears to be due at least in part to shifts in their rooting profile, whereby a greater proportion of root biomass is produced deeper in the profile (Lopes and Reynolds, 2011; Reynolds et al., 2008).
Another approach to uncovering useful variation in Ae. tauschii accessions is to develop a Nested Association Mapping population (NAM), a method described for maize by Yu et al. (2008). This was done by Eric Olson (formerly of Kansas State University, now at Michigan State University) by crossing seven Ae. tauschii accessions directly to the Kansas adapted wheat line KS05HW14, followed by two backcrosses and several generations of self-pollination. The resulting population of 420 lines (named the DNAM) is shown growing in Video 5.
Video 5. Dr. Patrick Byrne shows research plots of different D-genome wheat accessions that were crossed with an adapted wheat line from Kansas. Drought tolerance traits are being studied.
5. Useful genes from wild wheat relatives
Genes from Aegilops species and other wild relatives have been extremely important for improving a number of traits in wheat, especially disease resistance. Kishii (2019) has compiled genes that have been identified or transferred from Ae. tauschii to wheat. These includes genes for resistance to leaf rust, stem rust, stripe rust, powdery mildew, Septoria tritici, Septoria nodorum, tan spot, cyst nematode, root knot nematode, Hessian fly, greenbug, Russian wheat aphid, wheat curl mite, and soil-borne cereal mosaic virus. Another table in Kishii (2019) compiles genes from Aegilops species other than Ae. tauschii. This list includes genes for resistance to eyespot, leaf rust, stem rust, stripe rust, powdery mildew, cyst nematode, root knot nematode, and greenbug.
The Wheat Genetics Resource Center at Kansas State University has been a leader in collecting, conserving, and exploiting wild wheat relatives for crop improvement. Germplasm released by the Center includes breeding lines for disease and insect resistance, bread making quality, and other traits.
In the realm of abiotic stress tolerance, Placido et al. (2013) showed that a chromosome segment translocation from Agropyron elongatum provided wheat with the ability to maintain root growth under moisture stress conditions.
6. References
Dreccer MF, Borgognone MG, Ogbonnaya FC, Trethowan RM, Winter B. 2007. CIMMYT-selected derived synthetic bread wheats for rainfed environments: Yield evaluation in Mexico and Australia. Field Crops Research 100:218-228.
Gepts P. 2018. The domestication of our food crops. In: Chrispeels MJ, Gepts P (editors). Plants, Genes & Agriculture: Sustainability through Biotechnology. Oxford University Press, Cary, North Carolina.
Kishii M. 2019. An update of recent use of Aegilops species in wheat breeding. Frontiers in Plant Science 10:585.
Lage J, Trethowan RM. 2008. CIMMYT’s use of synthetic hexaploid wheat in breeding for adaptation to rainfed environments globally. Australian Journal of Agricultural Research. 59:461-469.
Lopes MS, Reynolds MP. 2011. Drought adaptive traits and wide adaptation in elite lines derived from resynthesized hexaploid wheat. Crop Science 51:1617-1626.
Marcussen T, Sandve SR, Heier L, Spannagl M, Pfeifer M, International Wheat Genome Sequencing Consortium, Jakobsen KS, Wulff BB, Steuernagel B, Mayer KF, Olsen OA. 2014. Ancient hybridizations among the ancestral genomes of bread wheat. Science 345:1250092.
McFadden ES, Sears ER. 1946. The origin of Triticum spelta and its free—threshing hexaploid relatives. Journal of Heredity. 37:81-89; 107-116.
Mujeeb-Kazi A, Hettel GP, (editors). 1995. Utilizing wild grass biodiversity in wheat improvement: 15 years of wide cross research at CIMMYT. CIMMYT Research Report No. 2. Mexico City, Mexico.
Ogbonnaya FC, Abdalla O, Mujeeb‐Kazi A, Kazi AG, Xu SS, Gosman N, Lagudah ES, Bonnett D, Sorrells ME, Tsujimoto H. 2013. Synthetic hexaploids: Harnessing species of the primary gene pool for wheat improvement. Plant Breeding Reviews 37:35-122.
Placido DF, Campbell MT, Folsom JJ, Cui X, Kruger GR, Baenziger PS, Walia H. 2013. Introgression of novel traits from a wild wheat relative improves drought adaptation in wheat. Plant Physiology 161:1806-1819.
Reynolds M, Dreccer F, Trethowan R. 2007. Drought-adaptive traits derived from wheat wild relatives and landraces. Journal of Experimental Botany 58:177-186.
Rosyara U, Kishii M, Payne T, Sansaloni CP, Singh RP, Braun H-J, Dreisigacker S. 2019. Genetic contribution of synthetic hexaploid wheat to CIMMYT’s spring wheat breeding germplasm. Scientific Reports 9:12355.
Trethowan RM, Mujeeb-Kazi A. 2008. Novel germplasm resources for improving environmental stress tolerance of hexaploid wheat. Crop Science 48:1255-1265.
WHEAT. 2014. Wheat: Vital grain of civilization and food security. 2013 Annual Report, CGIAR Research Program on Wheat, Mexico City, Mexico.
Yu JM, Holland JB, McMullen MD, Buckler ES. 2008. Genetic design and statistical power of nested association mapping in maize. Genetics 178:539-551.
7. Acknowledgments
Citation: Byrne P. 2020. Case Study: Wheat Domestication and Breeding. In: Volk GM, Byrne P (Eds.) Crop Wild Relatives in Genebanks. Fort Collins, Colorado: Colorado State University. Date accessed. Available from https://colostate.pressbooks.pub/cropwildrelatives/chapter/wheat-breeding-with-crop-wild-relatives/
This training module was made possible in part by funding from USDA-ARS, Colorado State University, IICA-PROCINORTE (procinorte.net), and the United States Agency for International Development (USAID).
Chapter editors: Emma Balunek, Katheryn Chen, Gayle Volk
Videographer: Mike May
Photo banner photo credits (left to right): Phil Westra, Pat Byrne, Mike May.
This project was funded in part by the National Academy of Sciences (NAS) and USAID, and any opinions, findings, conclusions, or recommendations expressed in such are those of the authors alone, and do not necessarily reflect the views of USAID or NAS.


